→ return to THE news archive
Budding yeast and proteomics defined protein N-terminal acetylation
First published at UiB.no December 22, 2022
Budding yeast is a useful model system in molecular biology. Here, researchers in Bergen and Ghent used it to define one of the most abundant protein modifications in eukaryotes: N-terminal acetylation.
Photo: Henriette Aksnes
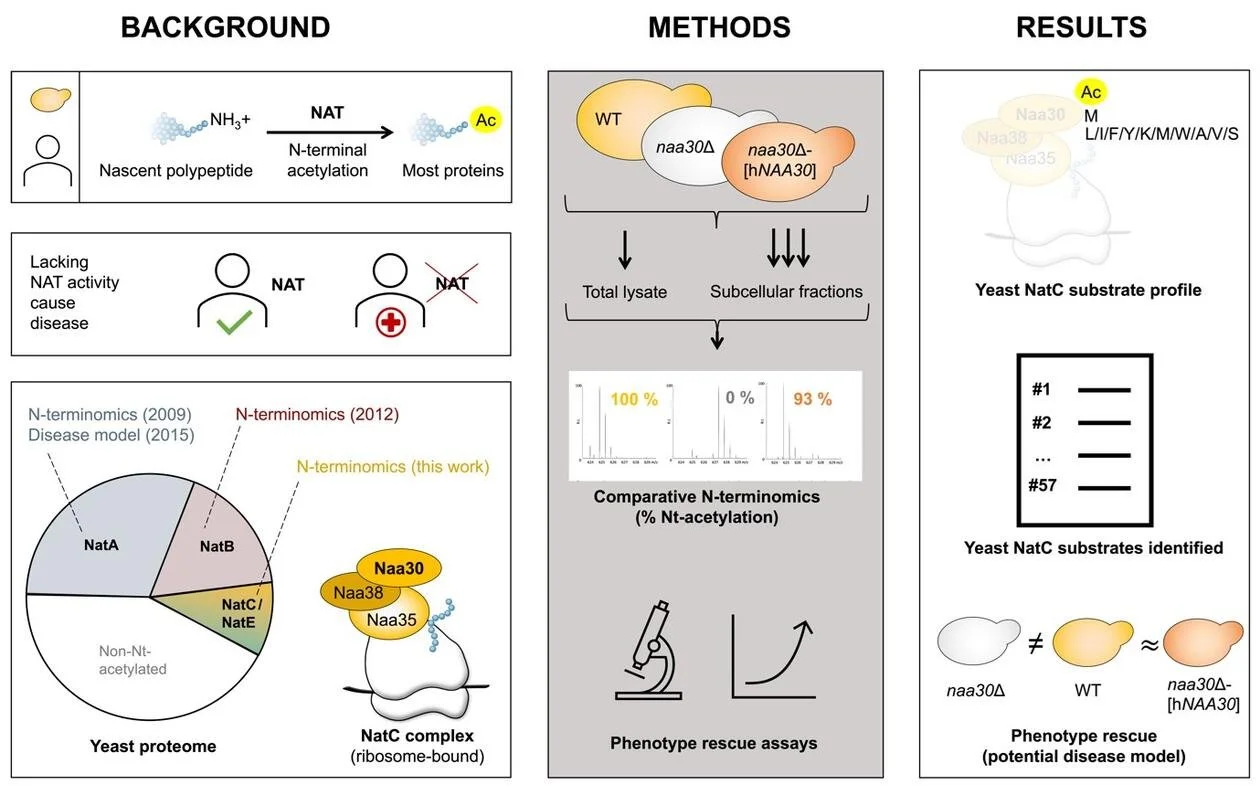
The yeast S. cerevisiae still serves as a valuable model for studying N-terminal acetylation. For two of the three major yeast N-terminal acetyltransferases (NatA and NatB) substrates were previously identified by N-terminomics. Together with our collaborators in Ghent, Arnesen lab now report the first yeast NatC-focused N-terminomics.
We identified 57 S. cerevisiae proteins N-terminally acetylated by NatC. Identification of a high number of substrates was possible through a yeast subcellular fractionation approach. We reveal here an expanded substrate profile for NatC compared to previous estimates. And we also found a hitherto unappreciated complex relationship between different yeast NATs.
This work offers increased functional insights into NatC-mediated N-terminal acetylation and provides a solid basis for future work to pinpoint the specific molecular mechanisms that link lack of NatC-mediated N-terminal acetylation to the described phenotypes of NatC-lacking yeast.
Pathogenic variants of genes encoding N-terminal acetyltransferases (NATs) have been and are continuously being uncovered. Our work underpins NatC-lacking yeast as a future disease model since we show that human NAA30 (catalytic subunit of NatC) expression in yeast can rescue naa30D yeast phenotypes and Nt-acetylation.
Expanded in vivo substrate profile of the yeast N-terminal acetyltransferase NatC
Petra Van Damme, Camilla Osberg, Veronique Jonckheere, Nina Glomnes, Kris Gevaert, Thomas Arnesen & Henriette Aksnes
J Biol Chem 2022. DOI: https://doi.org/10.1016/j.jbc.2022.102824. Online ahead of print.